Language and the Book of Life
The ambitious project of sequencing the DNA in the human genome released its first draft three years ago. At the time it was often portrayed as `reading the book of life’, but what is only recently being understood is how appropriate this metaphor really is. The jargon of molecular biology is scattered with terms borrowed from linguistics, such as transcribe, translate and code. The parallels between these two fields run much deeper than this though, and it has been discovered that both DNA and the proteins that it codes for have a grammatical structure like language. This has lead to the very productive swapping of ideas and techniques between biologists and linguists. For example, `authorship tests’ have been applied to unknown DNA sequences and `evolutionary trees’ have been constructed of old texts. But first, in what ways are DNA and proteins so profoundly similar to language?
The Structure of Language
Words within a sentence are by no means randomly ordered, but are strictly organised by the rules of grammar. This grammatical structure is hierarchical — much like a tree with layers of forked branches. Every sentence must contain a noun phrase and a verb phrase, each of which divides into further offshoots. The noun phrase for instance is composed of a determiner (such as the or a), some adjectives and the subjective noun. Languages differ slightly in their rules of syntax, for example in French the adjective usually comes after the noun and not before. Latin and the Star Wars character Yoda both swap the objective noun and verb branches to give sentences like `The girls hot toast like’. These are merely cosmetic differences though — no more than rotating two branches around each other — and all have the same fundamental tree–like construction.
Grammar also dictates that certain words in a sentence must agree with each other, such as the plurality of the subject noun and its verb. For example:
The girls like hot toast
is correct, whereas:
The girls likes hot toast
is not. A `dependency’ exists between the noun and its verb to ensure that they agree with each other.
The reason why language is so powerful and can generate an effectively unlimited number of different expressions is that it is also modular. This means that whole new phrase structures can be inserted into a sentence as subclauses:
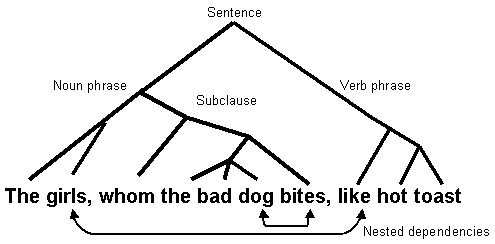
The girls, whom the bad dog bites, like hot toast
The crucial thing is that although girls and like have become separated in the sentence they must still agree with each other. This is known as a long-distance dependency, and is a defining feature of grammatical structures. The dependency stretches over the inserted subclause so that like agrees with girls and not dog, the noun nearest to it in the sentence. This leads to another important consequence. The subclause possesses its own dependency between dog and bites, which is contained within the long–distance dependency of the main sentence. The dependencies are said to be `nested’, as can be seen in the diagram below, which also shows the tree–like structure of grammar.
Grammar in proteins
Although grammatical structure is present in DNA it is more obvious in proteins and so we will concentrate on them. Long-distance dependencies are also present in proteins. Proteins are the workforce of a cell – they perform all the necessary jobs such as copying the DNA, transporting raw materials into the cell, or as enzymes catalysing the chemical reactions. They are made up of long chains of subunits called amino acids, which is directly equivalent to the string of words in a sentence. Every protein has a precise three-dimensional shape that allows it to perform its job, and so the chain must curl up in a very specific way and be held together by bonds between the amino acids. This folding means that amino acids a long distance from each other in the sequence end up being brought close together and bonding. Only certain amino acids bond with each other, and so for a protein to form correctly there exist rules that dictate which amino acids can go at certain positions in the chain. The result of this is that protein sequences possess long-distance dependencies in exactly the same way as sentences.
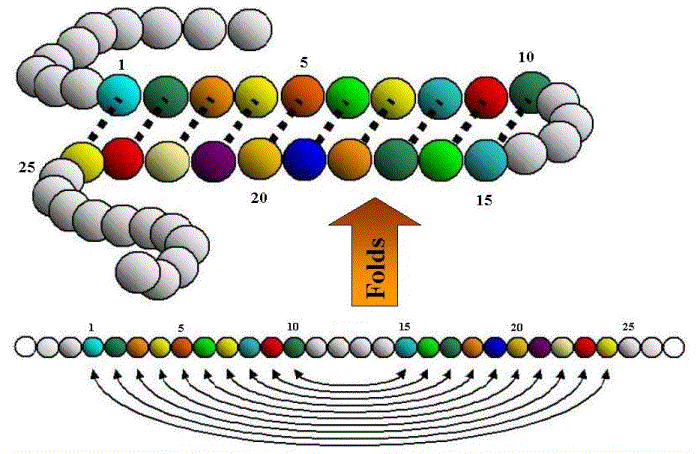
One common feature in proteins is known as an antiparallel sheet. This is produced when the amino acid chain bends back on itself several times, so that the strands run closely alongside each other, but in opposite directions (`antiparallel’). Fibroin, the protein in silk, is composed almost entirely of stacks of these antiparallel sheets. As can be seen below, this doubling-back leads to a series of nested dependencies, just like subclauses in a sentence:
There is another type of dependency but it is much less common than the nested one just described. The Dutch language is well known within linguistic circles for its very unusual ordering of words within certain subordinate clauses:
 Which translates to: (Bill knows) that John wants to let Mary read the book
Which translates to: (Bill knows) that John wants to let Mary read the book
The pattern of dependencies is very different in this situation. The connections are not nested as in the previous example, but leap-frog over each other. These are known as cross-serial dependencies, and are remarkably rare within natural languages. Although grammatically correct, they make a sentence hard to understand even if Dutch is your first language.
It appears that such cross-serial dependencies are also uncommon in proteins, as only a few examples have been discovered so far. The reason behind this is less clear than for languages, but it is believed to be down to folding. Proteins are manufactured in the cell as long chains of amino acids that must then spontaneously curl up into a precise arrangement. Certain structures, such as the antiparallel sheets described above, readily organise themselves and so are common among proteins. But structures that result from cross-serial dependencies may not fold as favourably and would be rare.
Meta-dependencies
Besides linking amino acids within the same chain, dependencies can also occur between different proteins. Many proteins are shaped so that they can interact with each other. For example, the machinery that copies DNA consists of at least four different proteins joined together by amino acid bonds between them. This means that dependencies link amino acids not only within the same chain, but also between completely separate proteins.
These meta-dependencies are important within literature as well. Often a writer will choose certain words to generate a coherent mood or adhere to an extended metaphor. Poetry is by far the best comparison, as word choice and order are limited not just by grammatical constraints, but also by consideration of the rhythm and rhyming scheme. Limericks have particularly strictly defined metres, with a rhyming pattern that is usually aabba. An example would be:
There was an old man from Peru,
Who dreamed he was eating his shoe.
He woke in a fright
In the dead of the night
And found it was perfectly true.
If the last word is swapped with correct, then both the syntax and meaning of the sentence would be unaltered since true and correct are synonyms. The rhythm and rhyming pattern of the limerick would be destroyed, however. There is a meta–dependency linking the words shoe and true that crosses over sentences. This is exactly like the grammatical links between amino acids that must stretch between separate proteins.
Semantic constraints
The parallels between language and protein sequences run even deeper than this. The function of language is to communicate information, and so as well as being grammatically correct a sentence should also be meaningful. Noam Chomsky, considered to be the father of modern linguistic analysis, perfectly illustrated this point with the example:
Colourless green ideas sleep furiously
With regards to syntax this is a faultlessly constructed sentence. Adjectives, nouns, verbs and adverbs all appear in the correct place in the sentence. Where necessary the words agree with each other, such as the dependency between ideas and sleep. It makes absolutely no sense, however, and so has no semantic value. The same is true of proteins. An amino acid sequence can be perfectly arranged with nested dependencies that rapidly fold into a well-formed protein. If it is not of a particular shape though, it will be unable to perform any job within the cell and so the protein is functionally useless, just like the above sentence. Many inheritable diseases, such as cystic fibrosis, are caused by a single mutation changing one of the amino acids in a protein. This disrupts the normal shape of the protein and it can no longer perform its job adequately. The semantic value of the protein has been lost as it cannot interact properly with other molecules.
A sentence may contain nonsensical words but still retain its meaning. Over a quarter of the words in Lewis Carroll’s popular poem Jabberwocky were invented, yet the sense of the piece is still largely intact. Semantics are derived from the words present and how they interact with each other, and so context can be enormously helpful in deducing the meaning of a previously unseen word. Purely from the background provided by recognised words it is obvious that a Jabberwock is a fearsome monster and a vorpal blade is probably a sharp one. In fact, galumphing has since been entered into the Oxford English dictionary as denoting a triumphant gait. This inference of function from the context is also becoming very important in molecular biology. Most of the domains, or functional areas, within a newly discovered protein may already be well known. Or at least if they are similar to known domains than it can usually be safely presumed that they perform similar jobs, in the same way that words with the same root often have related meanings. If a domain is completely novel, however, then its function could still be inferred from the context provided by other domains in the protein. For example, if a newly discovered protein has one domain that allows it to bind to DNA and another to a copying-regulation protein, then it would be reasonable to expect that the third, unknown, domain is involved with copying DNA.
There are fundamental parallels between language and molecular biology. Once identified they have triggered the productive exchange of ideas between linguists and biologists. Examples of these recent collaborations include applying prose authorship tests to DNA sequences, and drawing evolutionary trees of classic literary works.
Authorship tests
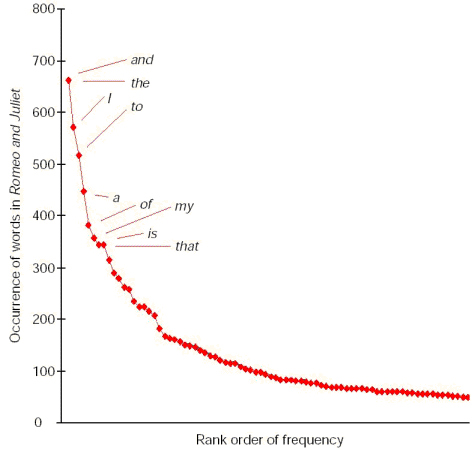
Textual analysis can be used to demonstrate the authenticity of disputed works. Each author has their own preference for using certain words, and so one technique compares the occurrence of different words in the uncertain text with that of an author’s known works. The counted words are ranked (whereby the most common is number one and the rarest is last) and then plotted on a graph with their frequency of occurrence up the side:
This is known as a Zipf graph, after the Hungarian linguist who first plotted one, and it clearly shows that a few common words account for a disproportionate amount of any text. Comparing the Zipf graphs of two different pieces of writing, paying attention to the position of selected words, reveals whether they were both composed by the same author.
The same shape of graph is found when aspects of DNA and proteins are plotted. The DNA code is made up of four letters – A, T, G and C – and certain short sequences of these letters turn up again and again in particular species but not in others. The authorship test can also be performed on an unfamiliar DNA fragment to discover which species it belongs to. Bacteria are infamous for being able to transfer sections of DNA between each other, which can transform a previously harmless bug into a highly contagious and deadly germ. The particular strain of E. Coli that is frequently in the news for causing outbreaks of food poisoning was created when these normally beneficial gut bacteria acquired virulence genes from another species. Microbiologists looked at the occurrence of certain short DNA sequences in these genes and were able to identify the characteristic signature of a known harmful bacterium, thus solving the riddle of how one strain of E. Coli became lethal.
Evolutionary Trees
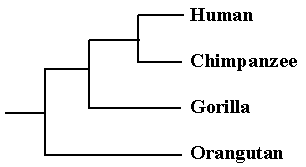
The adoption of literary techniques has enriched the field of molecular biology and helped resolve some long-running puzzles. The opposite is also true, and established genetic procedures have been applied to linguistic problems with great success. Evolutionary trees can be constructed on the basis of similarities in the DNA code of different species. Changes to the DNA letters, or mutations, are assumed to happen randomly, and so if two species contain only a few differences it is assumed that they share a recent ancestor. An evolutionary tree is then drawn with closely related species on neighbouring branches. For example, when the DNA of primates was analysed the results confirmed what zoologists had suspected for a long time. Humans were placed on the branch next to chimpanzees, with gorillas as evolutionary cousins, and orangutans as more distant relatives:
In 1998 biologists at Cambridge University applied exactly the same technique to analyse surviving versions of The Wife of Bath’s Prologue from Chaucer’s Canterbury Tales. During the fifteenth century scribes hand-copied this text many times and inevitably introduced mistakes and alterations so that each of the 58 versions is slightly different. These copying errors can be treated as mutations, and each of the variants as a different species. The number of differences between each version was counted and the most similar ones were placed next to each in a literary tree. Not only did this show the relations between different versions, but even allowed the scientists to backtrack down the tree to its trunk and calculate the `ancestral sequence’. They were able to reconstruct what the actual text of Chaucer’s original would probably have said, even though it has now been lost. The surprising thing is that in some parts the currently accepted version is significantly different from this reconstructed one.
The Future
The sequencing of DNA from various species is routinely generating enormous volumes of data that are simply impossible to analyse by hand. There is a huge demand for computer programs that can trawl through this raw information to find patterns and correlations that when investigated may prove to be important, a process known as data mining. One particularly exciting area is the development of algorithms that can spot previously unknown genes within DNA sequences, and this is where linguistic techniques are really making an impact.
Genes are often marked out in the DNA with special codes that tell the copying enzymes where they begin and end. The most basic gene-spotting algorithms simply search through the DNA sequence looking for these codes. It is rather like looking through a book containing mostly random letters and identifying anything that comes between a capital letter and a full stop as a sentence. The problem is that genes are rarely so clearly laid out and are often jumbled up, so would be overlooked by such an algorithm. The most advanced algorithms being designed today are based on techniques originally developed by linguists. They take into account the grammatical structure of proteins and genes described above and so are not fooled by disorganised genes.
The parallels between language and biological molecules are very deep. Both sentences and proteins possess a grammatical structure – shown here by long-distance dependencies. The reason behind this is not clear, but is probably due to the fact that they are both systems for conveying information, and so must be coherently structured. Both are also subject to the higher-level constraint that they must have semantic value and actually perform a set function. With such fundamental similarities between these two fields it is not surprising that researchers are increasingly starting to collaborate and exchange ideas and techniques. The adoption of new approaches is proving extremely successful and has already produced some very exciting results.
Language is one of the most ancient traits of humanity, and so it is perhaps apt that the techniques used to analyse it are becoming very important in one of our most modern exploits – that of studying our own genetic makeup, our book of life.
This article won Second Prize in the Times Higher Education & Oxford University Press science writing competition.